Histone deacetylase 1 (HDAC1; also known as GON-10, RPD3, or KDAC1) is a key epigenetic regulator involved in chromatin remodeling and gene expression. Aberrant HDAC1 activity has been implicated in tumorigenesis, and it is widely recognized as a high-value target for cancer therapy due to its role in promoting oncogenic transcriptional programs.
The mechanistic target of rapamycin (mTOR; also referred to as FRAP1, RAFT1, or RAPT1) is a central serine/threonine kinase that integrates nutrient and growth factor signals to regulate cell growth, proliferation, metabolism, and autophagy. Hyperactivation of mTOR signaling is a hallmark of various malignancies, contributing to uncontrolled tumor cell survival and resistance to therapy.
OTAVA’s Dual HDAC1/mTOR Inhibitors Library comprises 283 compounds with predicted dual inhibitory activity against both HDAC1 and mTOR. The library was developed using an advanced XGBoost machine learning algorithm trained on curated bioactivity datasets. Rigorous cheminformatics workflows — including structural filtering, Tanimoto-based clustering, and principal component analysis — were applied to ensure high structural diversity and optimal physicochemical properties suitable for drug discovery.
This library presents an opportunity to explore the synergistic inhibition of epigenetic and signaling pathways in cancer and beyond.
Target Relevance: HDAC1 and mTOR
-
HDAC1 regulates gene expression via histone deacetylation. Its inhibition can activate tumor suppressor genes and trigger apoptosis in cancer cells.
-
mTOR regulates cellular growth, metabolism, and angiogenesis. mTOR inhibitors are approved for cancer, autoimmune diseases, and post-transplant therapy.
-
Dual inhibition of HDAC1 and mTOR shows synergistic anticancer activity and the ability to overcome drug resistance. [1]
Library Preparation
1. Dataset preparation from ChEMBL
-
Activity data retrieved for HDAC1 and mTOR; duplicates and inconsistent structures removed.
-
Class labeling: active = pChEMBL > 5 (equivalent to IC₅₀/Ki < 10 µM), inactive = pChEMBL ≤ 5
2. Feature generation
-
Morgan fingerprints (radius = 2, 1024 bits)
-
Physicochemical descriptors: MolWt, LogP, TPSA, HBA, HBD
3. XGBoost model training
-
HDAC1: Accuracy = 0.92, ROC AUC = 0.97
-
mTOR: Accuracy = 0.96, ROC AUC = 0.99
4. Screening of OTAVA stock compounds
Novelty and Diversity Assessment
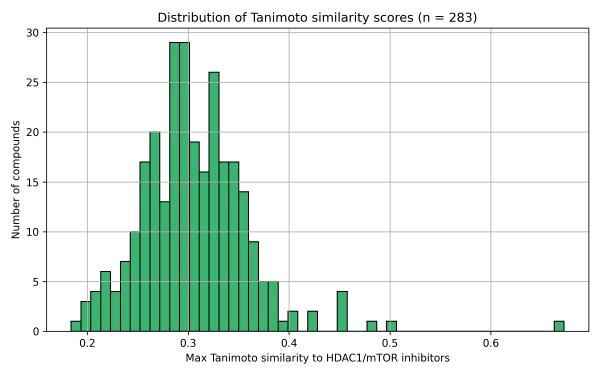
1. Tanimoto Similarity
2. Butina Clustering
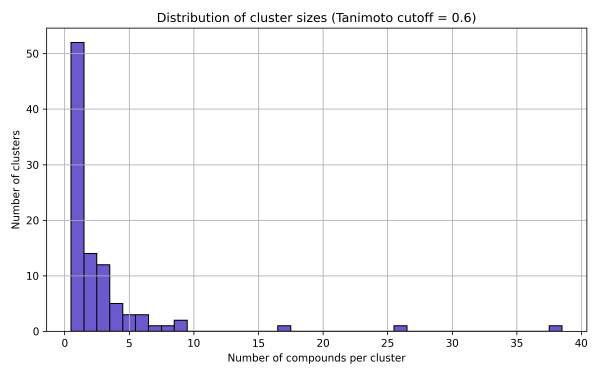
This result highlights a high level of structural diversity within the library. The presence of numerous singleton clusters indicates unique chemotypes, while the existence of one larger cluster may point to a promising core scaffold suitable for further SAR exploration or hit expansion.
3. PCA of Physicochemical Space
-
Descriptors: MolWt, LogP, TPSA, HBA, HBD
-
PC1 + PC2 explain over 77% of variance
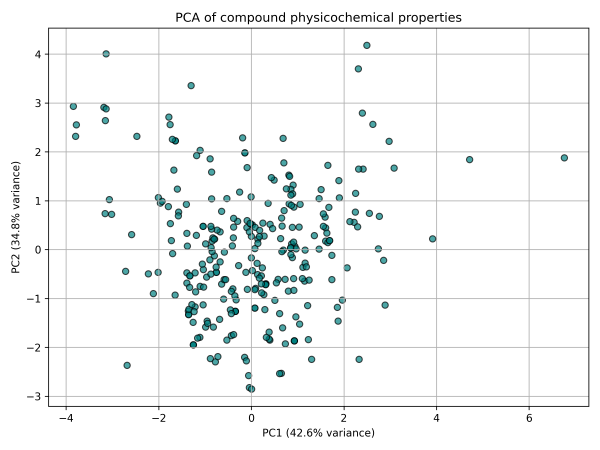
This demonstrates that the physicochemical space of the library is well-distributed and non-redundant. The compounds occupy diverse regions of drug-like chemical space, which increases the probability of finding hits with favorable ADME/Tox properties and target specificity.
4. PCA colored by Tanimoto clusters
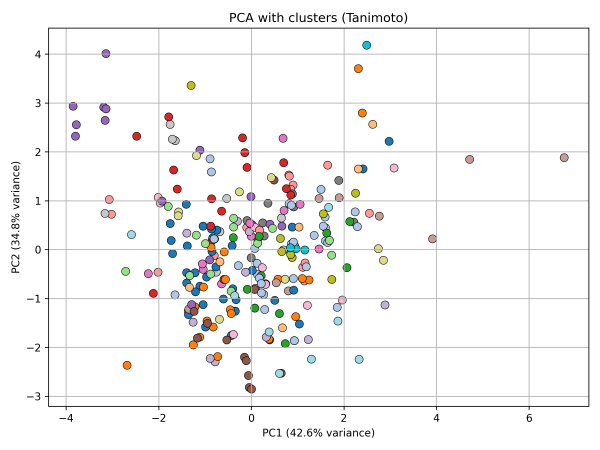
5. PCA highlighting structural outliers (Tanimoto < 0.25)

Applications of the Library
-
High-throughput screening for dual HDAC1/mTOR inhibitors
-
Hit identification and lead optimization
-
SAR exploration for dual-target scaffolds
-
Evaluation of synergistic inhibition strategies
Contact us today to discuss library access, customization, and collaborative screening strategies tailored to your research needs.
Key References
1. Yong Chen, Xue Yuan, Wanhua Zhang, Minghai Tang, Li Zheng, Fang Wang, Wei Yan, Shengyong Yang, Yuquan Wei, Jun He, Lijuan Chen. Discovery of Novel Dual Histone Deacetylase and Mammalian Target of Rapamycin Target Inhibitors as a Promising Strategy for Cancer Therapy. J. Med. Chem. 2019, Vol. 62(3), pp. 1577−1592, DOI: 10.1021/acs.jmedchem.8b01825.
 HOME
HOME ABOUT
ABOUT
 SERVICES
SERVICES
 PRODUCTS
PRODUCTS
 Targeted Libraries
Targeted Libraries
 Biochemicals
Biochemicals
 RESEARCH
RESEARCH
 DOWNLOADS
DOWNLOADS ORDERING
ORDERING
 CONTACTS
CONTACTS