|
SARS-CoV-2 NSP16 Targeted Library |
|
 2′-O-methyltransferase (2′-O-MTase, non-structural protein 16, nsp16) provides the viral mRNA with the ability to camouflage and obscure itself from the host cell, thus preventing recognition and activation of the host immune response which is essential for successful viral infection. Therefore, this protein can act as potential drug target for the SARS-CoV-2. 2′-O-methyltransferase (2′-O-MTase, non-structural protein 16, nsp16) provides the viral mRNA with the ability to camouflage and obscure itself from the host cell, thus preventing recognition and activation of the host immune response which is essential for successful viral infection. Therefore, this protein can act as potential drug target for the SARS-CoV-2.
OTAVAchemicals offers SARS-CoV-2 2′-O-methyltransferase (NSP16) Targeted Library, which contains 976 compounds with predicted activity against SARS-CoV-2 NSP16. The library has been designed with receptor-based virtual screening using crystal structure of SARS-CoV-2 2′-O-MTase (PDB ID: 6WKQ). The overall procedure included accurate flexible docking of Drug-like Green Collection into methyltransferase catalytic site. Final selection of compounds was made with inspection of enzyme active site’s crucial structural determinants for ligand binding, docking scores and intermolecular hydrogen bonds with key active site’s amino acid residues.
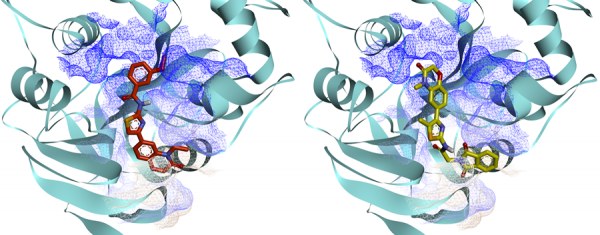
Example of complexes of ligands with NSP16,
obtained by molecular docking
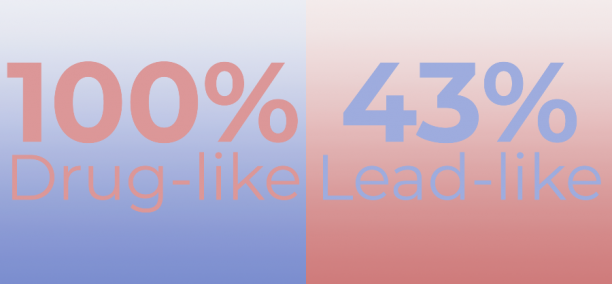
The designed SARS-CoV-2 NSP16 Targeted Library comprises only drug-like compounds (PAINS compounds are filtered off). The library is intended for screening projects to find new compounds with activity against SARS-CoV-2.
All compounds are in stock, cherry-picking is available.
The SARS-CoV-2 NSP16 Targeted Library (DB, SD, XLS, PDF format) as well as the price-list is available on request. Feel free to contact us or use on-line form below to send an inquiry if you are interested to obtain this library or if you need more information.
The summary of the SARS-CoV-2 NSP16 Targeted Library characteristics (average values):
|
Parameter |
Value |
|
MW |
394.7 |
|
ClogP |
3.3 |
|
ClogS |
-5.6 |
|
Number of Halogen Atoms |
0.4 |
|
Number of Rotatable Bonds |
4.3 |
|
Number of H Donors |
1.3 |
|
Number of H Acceptors |
4.5 |
|
PSA |
92.8 |
|
Number of Rings |
4.1 |
|
Number of Aromatic Rings |
3 |
|
Fraction of Sp3-Hybridized Carbons |
0.2 |
|
 HOME
HOME ABOUT
ABOUT
 SERVICES
SERVICES
 PRODUCTS
PRODUCTS
 Targeted Libraries
Targeted Libraries
 Biochemicals
Biochemicals
 RESEARCH
RESEARCH
 DOWNLOADS
DOWNLOADS ORDERING
ORDERING
 CONTACTS
CONTACTS