|
 The Retinoic Acid Receptor-related Orphan Receptor-γ (RORγ, RORc, NR1F3, RORG, RZR-GAMMA, RZRG, TOR or IMD42) plays an important role in metabolism, inimmunity and circadian rhythm. Abnormal functioning of RORγ is associated with inflammatory, immune and skin diseases including psoriasis. Antagonists and inverse agonists of this receptor may have therapeutic potential in the treatment of such diseases. The Retinoic Acid Receptor-related Orphan Receptor-γ (RORγ, RORc, NR1F3, RORG, RZR-GAMMA, RZRG, TOR or IMD42) plays an important role in metabolism, inimmunity and circadian rhythm. Abnormal functioning of RORγ is associated with inflammatory, immune and skin diseases including psoriasis. Antagonists and inverse agonists of this receptor may have therapeutic potential in the treatment of such diseases.
OTAVAchemicals offers Retinoic Acid Receptor-related (RAR-related) Orphan Receptor-γ Targeted Library. It contains 1174 predicted modulators of RORγ. Library has been carefully designed with combination of ligand-based and receptor-based virtual screening methods - machine learning (artificial neural networks, Bayesian statistics and k-nearest neighbors algorithm (k-NN)) and molecular docking.
For library design known RORγ antagonists and inverse agonists were clustered into four clusters. Compounds of each cluster were classified as active or inactive using k-NN based on molecular fingerprints to achieve more homogeneous sets and then randomly divided into training and test sets. The training sets were used for development and parameterization of artificial neural networks and for Bayesian modeling (both methods based on molecular descriptors, such as number of hydrogen donors and acceptors, PSA, LogP, molecular weight, number of rotatable bonds, number of rings and different fingerprints, topological descriptors and other). Also the training sets were used for compounds selection with k-NN algorithm (based on fingerprints ECFP4, FCFP4, ECFP6, FCFP6). The test sets were used for validation of neural networks and Bayesian models. Top-scored compounds obtained from application of these methods were crossed with top-scored compounds obtained with molecular docking (PDB ID: 4NIE). Final selection of compounds has been made with inspection of intermolecular hydrophobic contacts and hydrogen bonds with key ligand-binding domain residues. The combination of artificial neural networks, Bayesian statistics, k-NN algorithm and molecular docking should allow increasing the number of active compounds identified during screening.
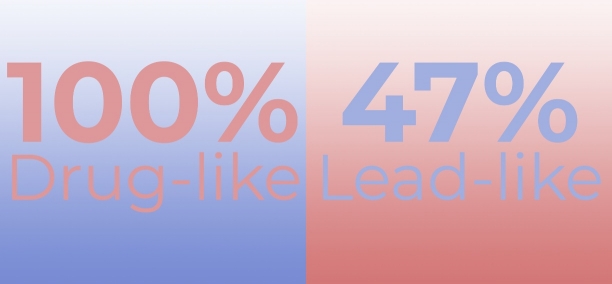
The designed RAR-related Orphan Receptor-γ Targeted Library comprises only drug-like compounds (PAINS compounds are filtered off). It provides an excellent basis for drug discovery projects related with the inflammatory, immune and skin diseases.
All compounds are in stock, cherry-picking is available.
The Retinoic Acid Receptor-related Orphan Receptor-γ Targeted Library (DB, SD, XLS, PDF format) as well as the price-list are available on request. Feel free to contact us or use on-line form below to send an inquiry if you are interested to obtain this library or if you need more information.
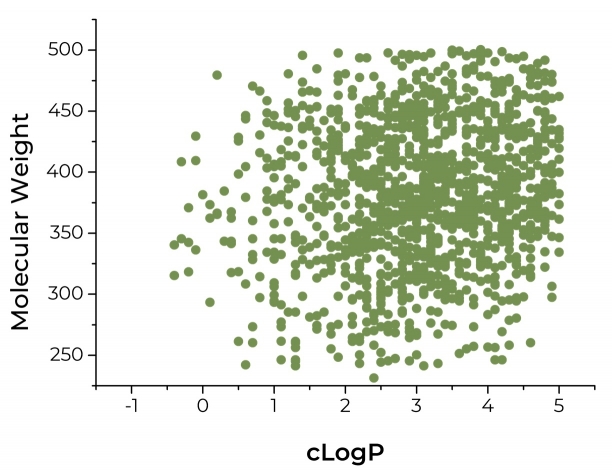
The summary of the RAR-related Orphan Receptor-γ Targeted Library characteristics:
|
Parameter |
Min |
Max |
Average |
|
MW |
217.3 |
500.4 |
380.5 |
|
ClogP |
-0.4 |
5 |
3 |
|
ClogS |
-8.1 |
-0.5 |
-4.9 |
|
Number of Halogen Atoms |
0 |
4 |
0.6 |
|
Number of Rotatable Bonds |
0 |
11 |
5 |
|
Number of H Donors |
0 |
4 |
1 |
|
Number of H Acceptors |
1 |
9 |
4.5 |
|
PSA |
29.1 |
172.6 |
84.9 |
|
Number of Rings |
2 |
6 |
3.4 |
|
Number of Aromatic Rings |
0 |
5 |
2.4 |
|
Fraction of Sp3-Hybridized Carbons |
0 |
0.97 |
0.3 |
|
 HOME
HOME ABOUT
ABOUT
 SERVICES
SERVICES
 PRODUCTS
PRODUCTS
 Targeted Libraries
Targeted Libraries
 Biochemicals
Biochemicals
 RESEARCH
RESEARCH
 DOWNLOADS
DOWNLOADS ORDERING
ORDERING
 CONTACTS
CONTACTS