|
We design ligand-based targeted libraries with powerful methods such as pharmacophore modeling, machine learning, QSAR or their combinations. Compound training sets are carefully created using data taken from ChEMBL, TIMBAL and other knowledge databases (Fig. 1).
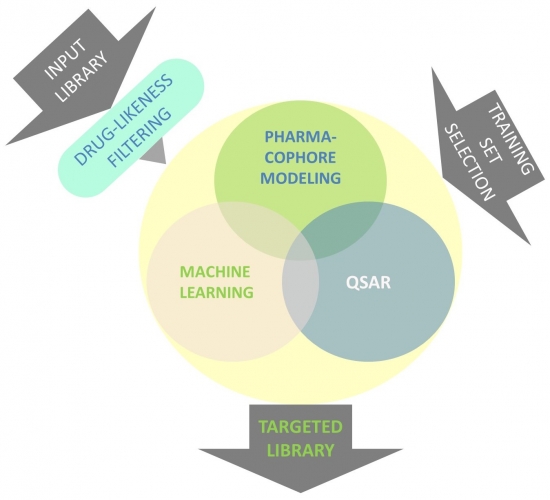
Fig. 1. Design flowchart of ligand-based targeted libraries.
Machine learning includes three powerful methods - artificial neural networks (NNET), Bayesian modeling and k-nearest neighbors algorithm (k-NN). General scheme of machine learning application is presented on figure 2.

Fig. 2. Application of machine learning for design of targeted libraries.
REFERENCES (PUBLISHED BY OTAVAchemicals SCIENTISTS)
-
Volynets GP, Starosyla SA, Rybak MYu, Bdzhola VG, Kovalenko OP, Vdovin VS, Yarmoluk SM, Tukalo MA (2019) Dual-targeted hit identification using pharmacophore screening. J Comput-Aided Mol Des 33:955-964.
-
Protopopov MV, Starosyla SA, Borovykov OV, Sapelkin VN, Bilokin YV, Bdzhola VG, Yarmoluk SM (2017) Hit identification of CK2 inhibitors by methods of virtual screening. Biopolym Cell 33(4):291-301.
-
Starosyla S.A., Volynets G.P., Protopopov M.V., Bdzhola V.G., Yarmoluk S.M. The development of algorithm for pharmacophore model optimization and rescoring of pharmacophore screening results. Ukrainica Bioorganica Acta, 2016, 14 (1), 24-34.
-
Starosyla SA, Volynets GP, Bdzhola VG, Golub AG, Protopopov MV, Yarmoluk SM. ASK1 pharmacophore model derived from diverse classes of inhibitors. Bioorg Med Chem Lett, 2014, 24 (18), 4418-4423.
-
Starosyla SA, Volynets GP, Bdzhola VG, Golub AG, Yarmoluk SM. Pharmacophore approaches in protein kinase inhibitors design. World J Pharmacol, 2014, 3 (4), 162-173.
-
Yakovenko et al. Kirchhoff atomic charges fitted to multipole moments: implementation for a virtual screening system. Journal of Computational Chemistry 2008, 29 (8), 1332-1343.
-
Yakovenko et al. The new method of distribution integrals evaluations for high throughput virtual screening. Ukrainica Bioorganica Acta 2007, 5 (1), 52-62.
-
Yakovenko et al. Application of distribution function of rotation and translation degrees of freedom for CK2 inhibitors Ki estimation. Ukrainica Bioorganica Acta, 2006, 4 (2), 47-55.
|
 HOME
HOME ABOUT
ABOUT
 SERVICES
SERVICES
 PRODUCTS
PRODUCTS
 Targeted Libraries
Targeted Libraries
 Biochemicals
Biochemicals
 RESEARCH
RESEARCH
 DOWNLOADS
DOWNLOADS ORDERING
ORDERING
 CONTACTS
CONTACTS



