|
 The pandemic of coronavirus disease 2019 (COVID-19), which is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a serious global concern for public health with thousands of fatalities. Today, no specific drugs are available to treat this disease. Thus, there remains an urgent need for the development of specific antiviral therapeutics toward SARS-CoV-2. The pandemic of coronavirus disease 2019 (COVID-19), which is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a serious global concern for public health with thousands of fatalities. Today, no specific drugs are available to treat this disease. Thus, there remains an urgent need for the development of specific antiviral therapeutics toward SARS-CoV-2.
OTAVAchemicals offers SARS-CoV-2 Targeted Libraries, which contain compounds with predicted activity against SARS-CoV-2:
-
SARS-CoV-2 Main Protease Targeted Library (1017 compounds);
-
ML (Machine Learning) SARS Targeted Library (1577 compounds).
The SARS-CoV-2 Main Protease Targeted Library has been designed with receptor-based virtual screening using crystal structure (PDB ID: 6LU7) of SARS-CoV-2 main protease (3C-like proteinase, 3CLpro, Mpro). The overall procedure included accurate flexible docking of Drug-like Green Collection into protease cleavage site. Final selection of compounds was made with inspection of enzyme active site’s crucial structural determinants for ligand binding, docking scores and intermolecular hydrogen bonds with key active site’s amino acid residues.
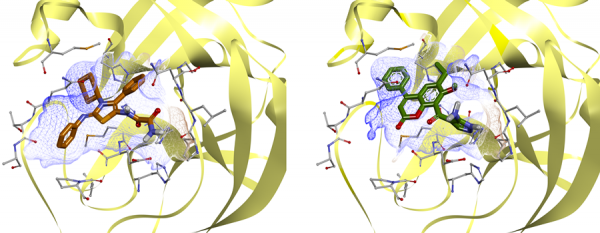
Example of complexes of potentially active ligands with main protease,
obtained by molecular docking
The ML SARS Targeted Library has been carefully designed using machine learning (artificial neural networks and Bayesian statistics).
For machine learning 306 compounds with anti-SARS activity were randomly divided into two equal groups. Each group had the same amounts of active and inactive compounds and was used as training and test set. The training sets were used for development of Bayesian and artificial neural networks models. Both methods were based on a number of different molecular descriptors - fingerprints, molecular weight, number of hydrogen acceptors and donors, number of rings, number of rotatable bonds, LogP, PSA, topological descriptors and other. The test sets were used for validation of all models.

Example of compounds from training set for machine learning and their anti-SARS activities
Virtual screenings of Drug-like Green Collection toward best Bayesian and artificial neural networks models were performed. Top-scored compounds, selected by machine learning methods, were visually analyzed. The application of number different artificial neural networks and models (21 in total) should allow increasing the number of active compounds identified during screenings.

Example of compounds selected by machine learning
The designed SARS-CoV-2 Targeted Libraries comprise only drug-like compounds (PAINS compounds are filtered off). The libraries are intended for screening projects to find new compounds with activity against SARS-CoV-2.
All compounds are in stock, cherry-picking is available.
The SARS-CoV-2 Targeted Libraries (DB, SD, XLS, PDF format) as well as the price-list are available on request. Feel free to contact us or use on-line form below to send an inquiry if you are interested to obtain these libraries or if you need more information.
The summary of the SARS-CoV-2 Targeted Libraries characteristics (average values):
|
Parameter |
SARS-CoV-2 Main Protease Targeted Library |
ML SARS Targeted Library |
|
MW |
408.8 |
349.1 |
|
ClogP |
3.8 |
3.3 |
|
ClogS |
-6.1 |
-4.9 |
|
Number of Halogen Atoms |
0.4 |
0.5 |
|
Number of Rotatable Bonds |
4.3 |
3.8 |
|
Number of H Donors |
1.1 |
0.8 |
|
Number of H Acceptors |
4.5 |
3.8 |
|
PSA |
88.6 |
75.7 |
|
Number of Rings |
4.4 |
6 |
|
Number of Aromatic Rings |
3.1 |
2.3 |
|
Fraction of Sp3-Hybridized Carbons |
0.2 |
0.3 |
|
 HOME
HOME ABOUT
ABOUT
 SERVICES
SERVICES
 PRODUCTS
PRODUCTS
 Targeted Libraries
Targeted Libraries
 Biochemicals
Biochemicals
 RESEARCH
RESEARCH
 DOWNLOADS
DOWNLOADS ORDERING
ORDERING
 CONTACTS
CONTACTS